Research
We are broadly interested in solving problems at the intersection of chemical biology, enzymology, bioorganic chemistry, and drug discovery (not necessarily all at the same time). Our research involves building holistic pipelines that integrate experimental methods with deep learning to develop original solutions to major contemporary challenges in chemical and pharmaceutical sciences. In addition to the summary below, check also our latest publications for a comprehensive overview of the recent lab activities.
A platform for ultra-high-throughput enzymology
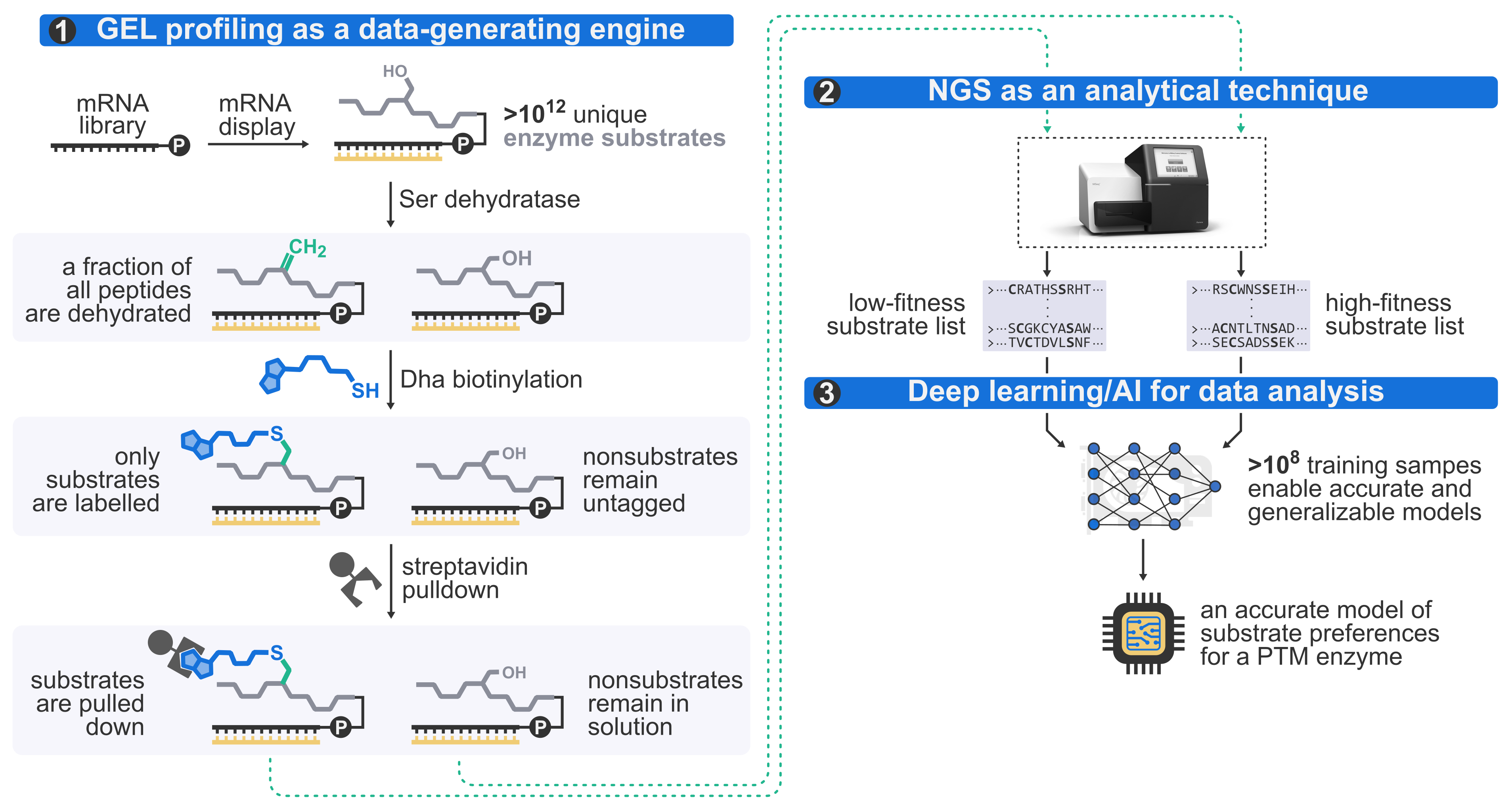
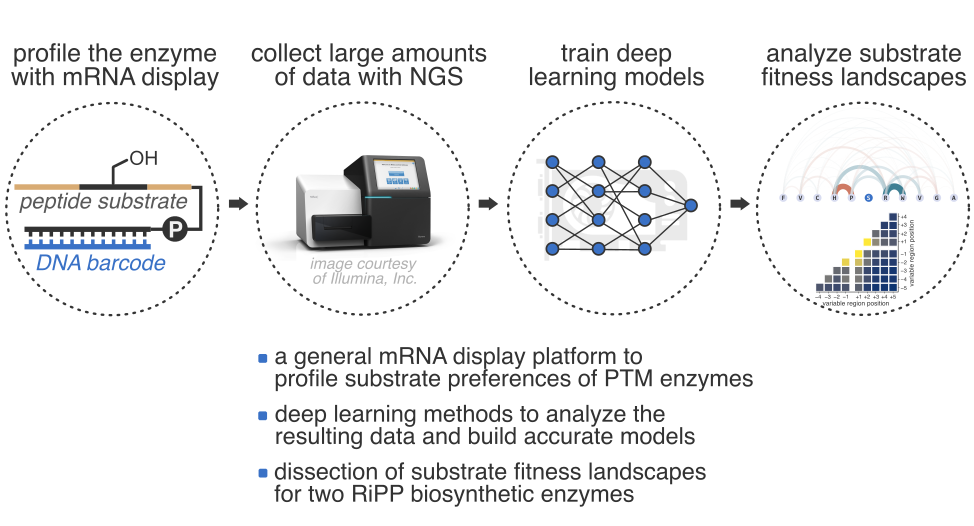
Enzymes are everywhere, and yet despite the decades of tremendous progress in fundamental enzymology, many aspects behind their remarkable catalytic efficiency remain poorly understood. We are building a general platform for ultra-high-throughput enzymology using genetically encoded libraries (mainly, mRNA display) to run a lot of enzymatic reactions at the same time (often as many as 10 trillion!), next generation sequencing to decode the reaction outcomes, and deep learning to analyze the data. The platform is a suite of experimental and computational tools to run massively parallel experiments in classical enzymology: e. g., one-shot measurement of millions of kcat/KM values or enzymatic kinetic isotopes effects; profiling trillions of enzymatic substrates all at once; and so on. Our aim with these is to harness the resulting data to furnish accurate, quantitative insights into the mechanistic aspects of enzymatic catalysis and elucidate the dynamics of the substrate engagement process. We focus on the questions which are either inaccessible or impractical using traditional techniques, and then use what we learn for engineering applications, e.g., as described in the section below.
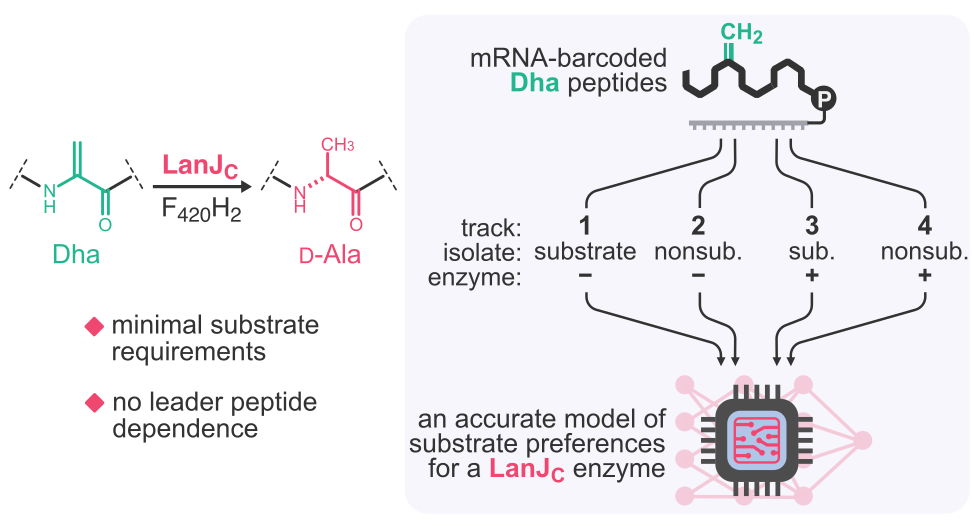
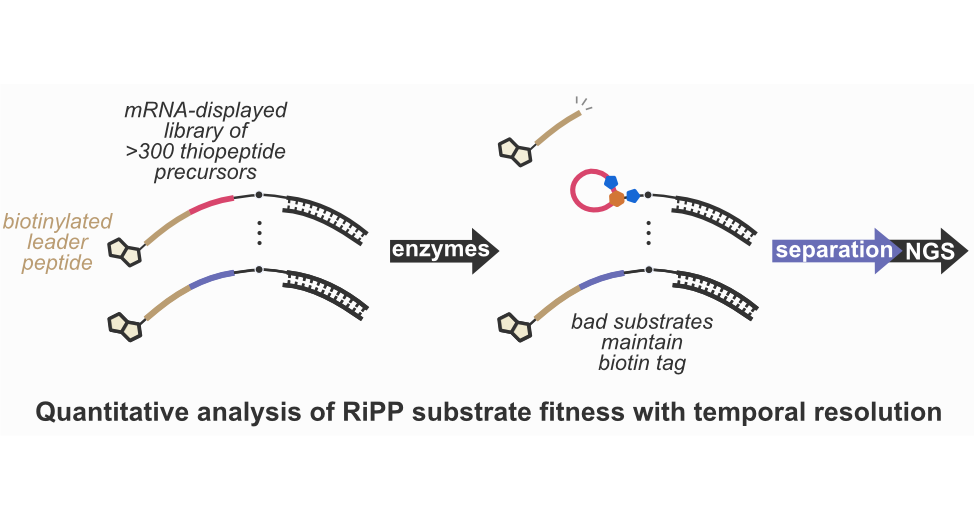
At present, much of our effort is dedicated to understanding catalysis by post-translational modification (PTM) enzymes, particularly the enzymes responsible for the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs), which is a large group of structurally diverse secondary metabolites with clinically important bioactivities. Many RiPP enzymes are highly catalytically promiscuous, i. e., capable of accepting millions of substrates, yet their substrate preferences are often highly nuanced and difficult to pinpoint using traditional batch experimentation. At present, there is no clear understanding of how such enzymes engage with and discriminate their substrates. We begin to address these questions with our platform by running rapid, systematic and unbiased profiling of the substrate fitness landscapes for promiscuous RiPP enzymes. The results unravel the complexity of the substrate engagement process and bring us closer to an atomic-level understanding. The “by-products” of these studies, deep learning models of enzymatic substrate preferences, become the foundation for our drug discovery pipelines.
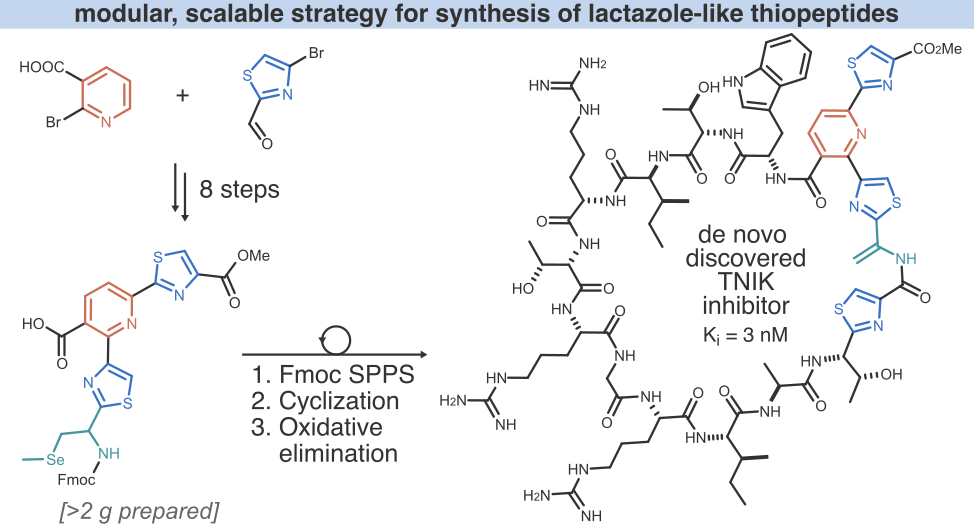
Natural product-like peptides in drug discovery
The use of natural products (NPs), i. e., secondary metabolites produced by bacteria, fungi and plants, in medicine goes back to prehistoric times. Despite the decades of impressive progress in drug discovery, NPs remain a major source of new pharmaceuticals — nearly half of all medications approved between 1981 and 2019 are NPs or their derivatives. This is not accidental; NPs evolved to mediate protein functions in complex biological systems, and their structures are selected for optimal stability, cell penetration, low off-target toxicity, and other pharmacological properties. Leveraging NP-like structures for the identification of lead compounds remains one of the most efficient strategies in early drug development.
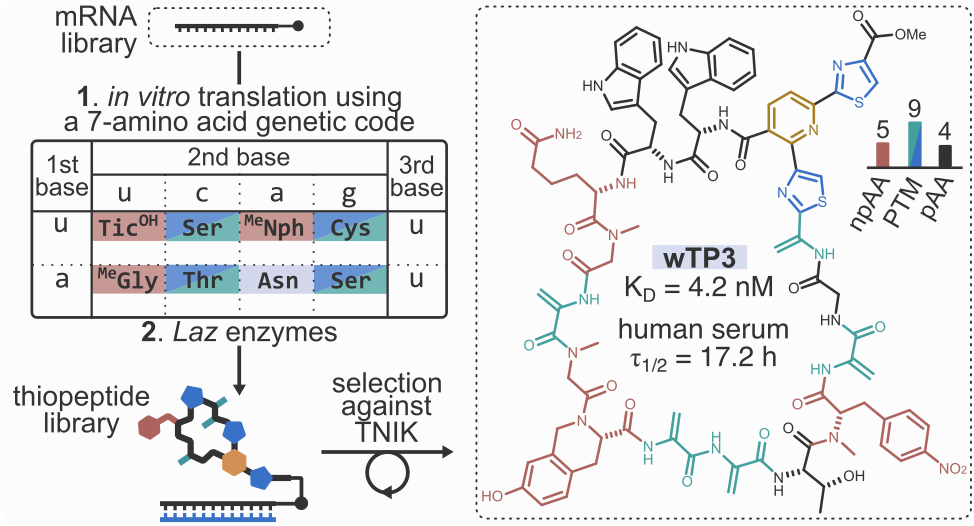
On the other hand, mRNA display-based discovery of macrocyclic peptides has also gained popularity in recent years. A number of mRNA display-derived compounds are currently undergoing clinical trials (see for example LUNA18 and MK-0616), and one peptide, zilucoplan, was recently approved by the FDA for the treatment of myasthenia gravis. These results have generated a sustained growing interest and renewed optimism in cyclic peptides as a drug modality. mRNA display can access ultra-large libraries (>1 trillion unique compounds) of macrocyclic peptides which enables rapid discovery of potent ligands against therapeutic targets of interest. However, as exemplified by the development of the aforementioned MK-0616, due to their often suboptimal pharmacological properties, mRNA display-derived peptides require extensive structural optimization before they can be considered as lead compounds.
To overcome the existing limitations of peptides as a drug modality, we combine the power of natural product biosynthesis with the throughput of mRNA display for rapid discovery of NP-like ligands with excellent pharmacological properties. We employ natural product biosynthetic enzymes to generate molecular diversity and leverage the deep learning models built using our enzymology platform to enable the biosynthesis in a combinatorial format.
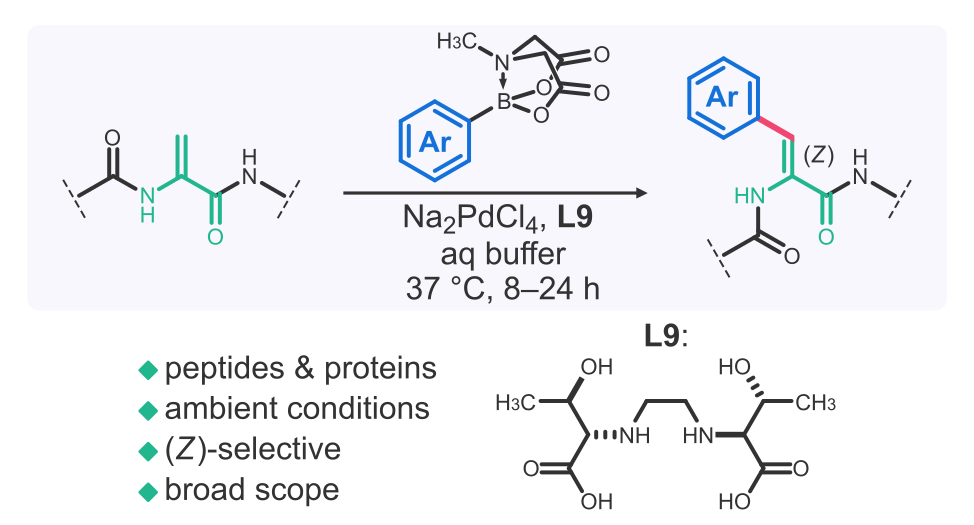
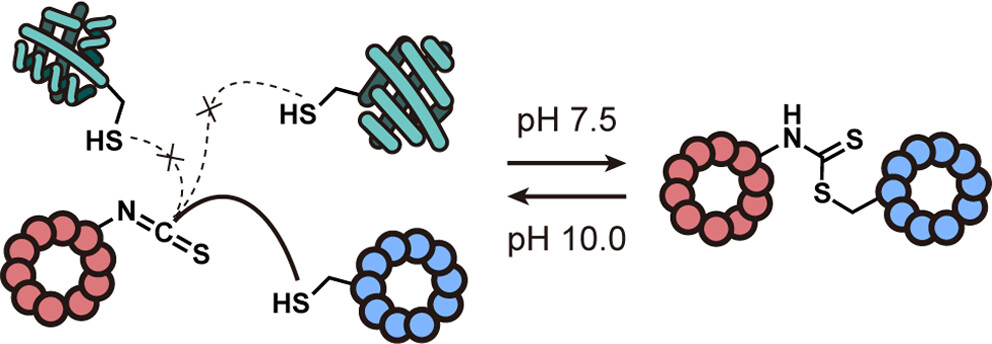
Late-stage modification of peptides and proteins
Late-stage modification of peptides is a powerful approach for generating molecular complexity and diversity. We develop methodologies to access structurally privileged peptides that are otherwise difficult to obtain, for use in drug discovery applications as outlined above. Backbone rigidification, macrocyclization, and side-chain preorganization are some of the structural features that we look to imbue our peptides with. We are problem-driven and employ both enzymatic and non-enzymatic approaches. Ultimately, we aim to develop chemistries that:
i) meaningfully improve peptide drug-likeness
ii) operate on fully unprotected peptides (and, by extension, proteins)
iii) proceed in water under ambient conditions (pH 4~9, 4~37 °C)
iv) have high chemo-, regio-, and stereoselectivity
v) have fast kinetics where possible
vi) can be integrated with modern drug discovery techniques like mRNA display